Figure 2
An unusual clonal chromosome abnormality der(17)t(11;17)(q24;p13)inv(11)(q13;q23) in a patient with chronic lymphocytic leukemia
Valentina Monti*, Fabio Serpenti, Lucia Farina, Maria Luisa Moiraghi, Maria Adele Testi and Giancarlo Pruneri
Published: 10 November, 2021 | Volume 5 - Issue 2 | Pages: 028-031
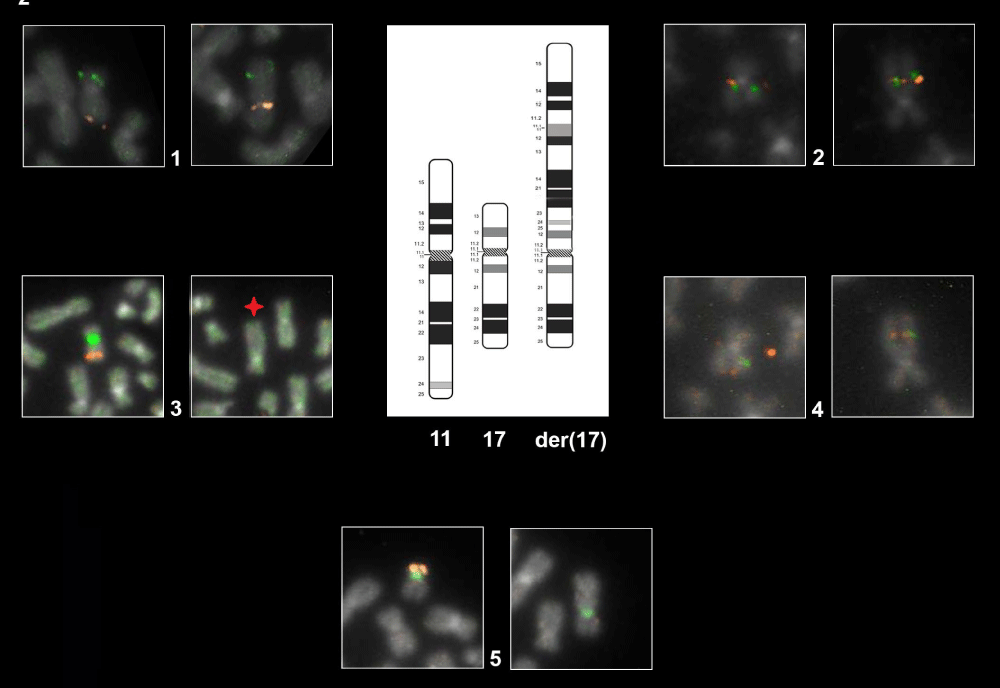
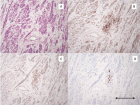
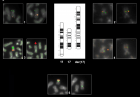
Figure 2:
Representative FISH Panel probes. The first eight cells each with two boxes marked with the number 1-2-3-4 show the normal ch.11 (left box) and the der(17) (right box) hybridized with different probes as follow: Box 1: WT1/FLI1 by combination probe set. FISH result highlight the der(17) orientation: green signal (WT1) map on the short arm of chromosome 11 and orange signal (FLI1) map on the long arm of chromosome 11. Box 2: CCND1 Dual Color Break Apart Probe Rearrangement. Der(17) harboring the CCND1 gene in closed proximity to 17p compared with normal chromosome. Box 3 LSI ATM/CEP11. As shown on normal ch.11 (left box) the orange fluorochrome hybridizes ATM gene and the green fluorochrome direct labeled the CEP11. The star on der(17) (right box) show the interstitial deletion affecting the gene ATM and the loss of CEP11. Box 4: MLL Dual Color Break Apart rearrangement. Der(17) harboring the MLL gene in closed proximity to 11p. The last two boxes, marked with the number 5, shows the normal ch.17 (left box) and der(17) in the (right box) hybridized with TP53 (orange)/CEP17(green). As illustrated der(17) keeps intact the CEP17.
Read Full Article HTML DOI: 10.29328/journal.acr.1001053 Cite this Article Read Full Article PDF
More Images
Similar Articles
-
An unusual clonal chromosome abnormality der(17)t(11;17)(q24;p13)inv(11)(q13;q23) in a patient with chronic lymphocytic leukemiaValentina Monti*,Fabio Serpenti,Lucia Farina,Maria Luisa Moiraghi,Maria Adele Testi,Giancarlo Pruneri. An unusual clonal chromosome abnormality der(17)t(11;17)(q24;p13)inv(11)(q13;q23) in a patient with chronic lymphocytic leukemia. . 2021 doi: 10.29328/journal.acr.1001053; 5: 028-031
Recently Viewed
-
Examining the Effects of High Poverty and Unemployment on Rural Urban Migration in Nigeria and its Consequences on Urban Resources and Rural DeclineTochukwu S Ezeudu*, Bilyaminu Tukur. Examining the Effects of High Poverty and Unemployment on Rural Urban Migration in Nigeria and its Consequences on Urban Resources and Rural Decline. J Child Adult Vaccines Immunol. 2024: doi: 10.29328/journal.jcavi.1001012; 8: 001-013
-
Nasal cytology in patients with previous SARS-CoV-2 infection: occurrence of atypical lymphocytesArturo Armone Caruso*, Anna Miglietta, Giovanni De Rossi, Liliana Nappi, Veronica Viola, Stefano De Rossi, Salvatore Del Prete, Clara Imperatore, Sabato Leo, Daniele Naviglio, Monica Gallo, Daniela Marasco, Lucia Grumetto. Nasal cytology in patients with previous SARS-CoV-2 infection: occurrence of atypical lymphocytes. Adv Treat ENT Disord. 2023: doi: 10.29328/journal.ated.1001014; 7: 001-006
-
Senile CataractRagni Kumari*. Senile Cataract. J Community Med Health Solut. 2024: doi: 10.29328/journal.jcmhs.1001041; 5: 001-007
-
Lecture: “First Aid to the Population in Case of Heat and Sunstroke during Accidents, Catastrophes, Natural Disasters and Terrorist Attacks” of the Subject “Life Safety” for Humanitarian and Technical UniversitiesShapovalov KA1,2*, Shapovalova LA. Lecture: “First Aid to the Population in Case of Heat and Sunstroke during Accidents, Catastrophes, Natural Disasters and Terrorist Attacks” of the Subject “Life Safety” for Humanitarian and Technical Universities. J Community Med Health Solut. 2024: doi: 10.29328/journal.jcmhs.1001042; 5: 008-014
-
Smart Cities and Aging Well: Exploring the Links between Technological Models and Social Models for Promoting Daily Social Interaction for Geriatric CareJocelyne Kiss*, Miguel A Reyes, James Hutson. Smart Cities and Aging Well: Exploring the Links between Technological Models and Social Models for Promoting Daily Social Interaction for Geriatric Care. J Community Med Health Solut. 2024: doi: 10.29328/journal.jcmhs.1001043; 5: 015-022
Most Viewed
-
Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth EnhancersH Pérez-Aguilar*, M Lacruz-Asaro, F Arán-Ais. Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth Enhancers. J Plant Sci Phytopathol. 2023 doi: 10.29328/journal.jpsp.1001104; 7: 042-047
-
Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case PresentationJulian A Purrinos*, Ramzi Younis. Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case Presentation. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001099; 8: 075-077
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Pediatric Dysgerminoma: Unveiling a Rare Ovarian TumorFaten Limaiem*, Khalil Saffar, Ahmed Halouani. Pediatric Dysgerminoma: Unveiling a Rare Ovarian Tumor. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001087; 8: 010-013
-
Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative reviewKhashayar Maroufi*. Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative review. J Sports Med Ther. 2021 doi: 10.29328/journal.jsmt.1001051; 6: 001-007

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."